How do we evolve with our natural enemies?
 The mushroom-feeding species Drosophila innubila
The mushroom-feeding species Drosophila innubila
“Now here, you see, it takes all the running you can do, to keep in the same place,” - Lewis Carroll
Life is rough. You've got to protect yourself against infection by pathogens and other natural enemies. That can include stuff from all walks of life: parasites, viruses, bacteria, fungi. Exactly what selective pressure you're facing varies over time: there might be a surge in a specific pathogen at a specific time in society (e.g. a pandemic event). It can also vary in geography, in location: not all places even have the introduction of the pathogen, or to the same extent. That requires hosts to adapt to novel threats arising over time, and how much they adapt varies from location to location. This doesn't just affect humans, but all animals face the same pressures. That's why we're at a loss in predicting or otherwise understanding which species will be threatened by a new zoonotic spillover (e.g. Chytridiomycosis, SARS-CoV-2). We know these principles of host-pathogen ecology, and we can list specific existing case studies with fascinating host-pathogen biological mechanisms. But when we talk about the broad rules of what makes a host susceptible or resistant to a given pathogen, we can't say much.
Insects and arthropods are incredibly important disease vectors - we can all name a list of things we'd prefer not to bite us (e.g. mosquitoes, sandflies, kissing bugs). Heck, the World Health Organization suggests that dengue-vector mosquitoes alone pose a risk to ~3.9 billion people globally. Insects are also major vectors of plant diseases. So it's very important to understand how insect immune systems work to understand how they are able to vector those diseases. We also want to control insects, and one way we're doing that is using natural enemies like viruses, fungi, and insect-parasites to suppress pest insect populations like aphids, without harming beneficial insects like bees. All of these biocontrol strategies will only be successful if we know how the parasite or pathogen works, and how the insect immune system responds.
Drosophila melanogaster has been an incredible model organism for understanding how insect immunity works. Drosophila genetics are in a class of their own - that's not just hyperbole, it's really very true. That makes Drosophila a great system to study insect immunity. However D. melanogaster is just one species, and it's not clear how representative it is of other insects. We don't even really know how representative it is of other Drosophila. My Wellcome-funded research seeks to inform on how immune processes in a model organism translate across species. For this, I use the genetic powerhouse Drosophila melanogaster and compare it to other Drosophila species. Each species has a unique ecology, and this will change the infectious pressures faced by the host. That makes fruit flies a fantastic model system, because we can rear literal tons of flies in a relatively small lab space, and study how they behave the same, or different, from D. melanogaster. This is the sort of work you simply couldn't do in other animals, unless you wanted to rear elephants, deer, mice, chimps, rhinos, bats, and koalas all in the same place - and even then, unlike with fruit flies, I'd probably feel bad having to infect them.
Life is rough. You've got to protect yourself against infection by pathogens and other natural enemies. That can include stuff from all walks of life: parasites, viruses, bacteria, fungi. Exactly what selective pressure you're facing varies over time: there might be a surge in a specific pathogen at a specific time in society (e.g. a pandemic event). It can also vary in geography, in location: not all places even have the introduction of the pathogen, or to the same extent. That requires hosts to adapt to novel threats arising over time, and how much they adapt varies from location to location. This doesn't just affect humans, but all animals face the same pressures. That's why we're at a loss in predicting or otherwise understanding which species will be threatened by a new zoonotic spillover (e.g. Chytridiomycosis, SARS-CoV-2). We know these principles of host-pathogen ecology, and we can list specific existing case studies with fascinating host-pathogen biological mechanisms. But when we talk about the broad rules of what makes a host susceptible or resistant to a given pathogen, we can't say much.
Insects and arthropods are incredibly important disease vectors - we can all name a list of things we'd prefer not to bite us (e.g. mosquitoes, sandflies, kissing bugs). Heck, the World Health Organization suggests that dengue-vector mosquitoes alone pose a risk to ~3.9 billion people globally. Insects are also major vectors of plant diseases. So it's very important to understand how insect immune systems work to understand how they are able to vector those diseases. We also want to control insects, and one way we're doing that is using natural enemies like viruses, fungi, and insect-parasites to suppress pest insect populations like aphids, without harming beneficial insects like bees. All of these biocontrol strategies will only be successful if we know how the parasite or pathogen works, and how the insect immune system responds.
Drosophila melanogaster has been an incredible model organism for understanding how insect immunity works. Drosophila genetics are in a class of their own - that's not just hyperbole, it's really very true. That makes Drosophila a great system to study insect immunity. However D. melanogaster is just one species, and it's not clear how representative it is of other insects. We don't even really know how representative it is of other Drosophila. My Wellcome-funded research seeks to inform on how immune processes in a model organism translate across species. For this, I use the genetic powerhouse Drosophila melanogaster and compare it to other Drosophila species. Each species has a unique ecology, and this will change the infectious pressures faced by the host. That makes fruit flies a fantastic model system, because we can rear literal tons of flies in a relatively small lab space, and study how they behave the same, or different, from D. melanogaster. This is the sort of work you simply couldn't do in other animals, unless you wanted to rear elephants, deer, mice, chimps, rhinos, bats, and koalas all in the same place - and even then, unlike with fruit flies, I'd probably feel bad having to infect them.
Antimicrobial peptides: natural antibiotics of the innate immune system
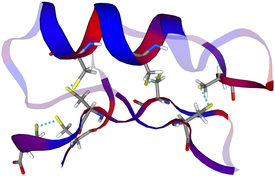 A 3D model of the AMP "Drosomycin"
A 3D model of the AMP "Drosomycin"
"We shape our buildings; thereafter they shape us." - Winston Churchill
Antimicrobial peptides (AMPs) are host-encoded antibiotics that combat invading microorganisms. These peptides are produced at pretty much any surface that comes in contact with microbes in the environment: in the gut, genitalia, skin, and at mucosal membranes in the lungs, eyes etc... My thesis work in Bruno Lemaitre's lab has shown that the constant secretion of these peptides is a key determinant in the composition of the microbiome, and in many host-microbe interactions. Their production is regulated by the "innate immune system," a rapid front-line defence against infection before the involvement of antigens and antibodies, i.e. before the "adaptive immune system." In vertebrates, AMPs are also known to regulate inflammatory signalling and other infectious processes. Dysregulation of AMPs is implicated in a number of diseases including susceptibility to chronic infections (e.g. atopic dermatitis), autoimmune disorders (e.g. inflammatory bowel disease), lung infections (e.g. in parkinson's disease patients), and neurodegenerative disorders (e.g. alzheimer's disease). Each of these processes can be impacted by intimate associations between hosts and their microbes.
AMPs must strike a fine balance: being toxic enough to kill invading microbes but not so toxic as to disrupt host cells or beneficial microbes of the gut microbiome. I use Drosophila as a model to understand how variation in AMP sequence can impact function, and how sequence evolution responds to infectious pressures. I've found that AMPs can have remarkably specific interactions with microbes, wherein a single AMP gene can dictate success or failure in fighting off infection. Over the course of fly evolution, species with highly-specialized ecologies also show microbe-specific patterns of antimicrobial peptide evolution. It's no exaggeration to say that this model in fruit flies teaches us that our resident microbes shape the immune repertoire present in our genomes. Through our ecology and behaviours, we expose ourselves to a common set of microbes over our lifetimes, and it turns out those microbes have also put pressure on our genomes to be prepared. We shape our buildings, thereafter they shape us. This research brings a number of interesting questions to the fore, regarding chronic infectious or dysbiosis syndromes such as inflammatory bowel, atopic dermatitis, chronic lung infections, etc... Could such chronic infectious diseases be treated if we could identify specific AMPs/effectors that are evolutionarily adapted to control them?
Antimicrobial peptides (AMPs) are host-encoded antibiotics that combat invading microorganisms. These peptides are produced at pretty much any surface that comes in contact with microbes in the environment: in the gut, genitalia, skin, and at mucosal membranes in the lungs, eyes etc... My thesis work in Bruno Lemaitre's lab has shown that the constant secretion of these peptides is a key determinant in the composition of the microbiome, and in many host-microbe interactions. Their production is regulated by the "innate immune system," a rapid front-line defence against infection before the involvement of antigens and antibodies, i.e. before the "adaptive immune system." In vertebrates, AMPs are also known to regulate inflammatory signalling and other infectious processes. Dysregulation of AMPs is implicated in a number of diseases including susceptibility to chronic infections (e.g. atopic dermatitis), autoimmune disorders (e.g. inflammatory bowel disease), lung infections (e.g. in parkinson's disease patients), and neurodegenerative disorders (e.g. alzheimer's disease). Each of these processes can be impacted by intimate associations between hosts and their microbes.
AMPs must strike a fine balance: being toxic enough to kill invading microbes but not so toxic as to disrupt host cells or beneficial microbes of the gut microbiome. I use Drosophila as a model to understand how variation in AMP sequence can impact function, and how sequence evolution responds to infectious pressures. I've found that AMPs can have remarkably specific interactions with microbes, wherein a single AMP gene can dictate success or failure in fighting off infection. Over the course of fly evolution, species with highly-specialized ecologies also show microbe-specific patterns of antimicrobial peptide evolution. It's no exaggeration to say that this model in fruit flies teaches us that our resident microbes shape the immune repertoire present in our genomes. Through our ecology and behaviours, we expose ourselves to a common set of microbes over our lifetimes, and it turns out those microbes have also put pressure on our genomes to be prepared. We shape our buildings, thereafter they shape us. This research brings a number of interesting questions to the fore, regarding chronic infectious or dysbiosis syndromes such as inflammatory bowel, atopic dermatitis, chronic lung infections, etc... Could such chronic infectious diseases be treated if we could identify specific AMPs/effectors that are evolutionarily adapted to control them?
The strain on scientific publishing
|
|
Scientists, whether as authors, editors, or reviewers, are overwhelmed by the dynamics of "publish or perish" that are promoted by current funding policies and decision-making biases. What's more, Plan S, the push for unfettered Open Access publishing, has unintentionally tied the revenue a journal receives for editorial work to the total output of articles, rather than the quality of those articles. As a result, many publishers can be motivated to publish as many papers as possible, without constraints imposed by a need for minimum quality.
Myself and my colleagues have been vocal about this issue since ~2020, and we wrote a paper on this topic. You can find the website covering that work here: https://the-strain-on-scientific-publishing.github.io/website/ |
