|
Drosophila geneticists benefit from decades of research and development, providing tools that can tackle literally any gene in the genome. At the click of your mouse, you can readily order flies that will express dsRNA to knock down your gene of interest for <$20 plus the cost of shipping. Most genes have putative mutant alleles disrupting gene expression or structure. Now, in the age of CRISPR, it's even easier to fill in the gaps where existing toolkits are less robust. But CRISPR involves capitol. Time and money, and energy. And after that, there is still a chance that your "good" mutation isn't as good as you thought it was. Take it from someone who's experienced this firsthand on more than one occasion: and I haven't even worked with that many flies generated using CRISPR and double gRNA! Briefly: we generated and tracked a Cecropin mutation using mutant-specific primers, but somehow failed to detect a wild-type Cecropin locus present in our Cecropin-mutant stocks caused by a bizarre recombination event. More recently, I detected that a similar double gRNA approach that also included an HDR vector (two chromosome arms that were there as guides to the locus) did not insert as one might expect... I detected this after performing maybe the 3rd or 4th routine-check PCR where I eventually realized: "hey... that band is ~100 bp larger than it should be... isn't it?" In the end, instead of replacing the locus, the HDR vector failed to do its job and inserted in the middle of the promoter. SNPs and a 10 nt indel prevented detection by existing primers for the wild-type gene. In the end, our mutant was still effectively a hypomorph (which is good, because it had a phenotype we spent a good while on), but not a full knock out (Fig 1). 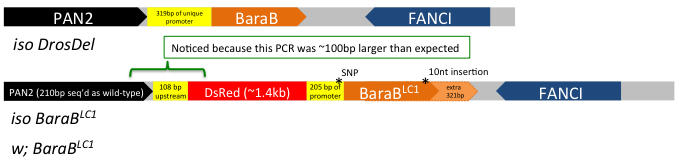 Figure 1: the BaraB[LC1] mutation (Hanson and Lemaitre, 2021) did not insert as expected. This mutation disrupts the promoter but the full gene remains present and mostly unchanged. A 10nt insertion at the C-terminus overlapped the reverse primer binding site that was initially used to check for the wild-type locus. My point here is that CRISPR isn't a magic tool that always works. Beyond these bizarre instances, off-target effects are a serious concern and could affect 4% of CRISPR mutant stocks (Shu Kondo, EDRC 2019); or more, it's tough to say! In total, CRISPR is a powerful tool in the toolkit, but it requires gRNA generation, injection, mutant isolation, and in my experience something that is especially important: robust mutant validation. This process takes a couple months at best, but more likely longer. But what if you didn't have to make new mutations? What if there was already an existing mutation you could access, but not in known databases. Enter the DGRP |
AuthorMark Archives
July 2024
Categories |
 RSS Feed
RSS Feed
