|
I was motivated to pen some thoughts on Furins owing the ongoing Covid-19 pandemic. My research focuses on antimicrobial peptides (AMPs), which in Drosophila and other insects are commonly cleaved by Furin-like enzymes. Relatively little has been done to understand exactly how different Furins act, and what intracellular processes they can regulate; this is likely at least in part due to their potential to affect many cell processes simultaneously, making genetic approaches difficult and confusticating the interpretation of focused in vitro approaches. What is Furin, and how does it relate to Sars-CoV-2? The Sars-CoV-2 (Sars2) pandemic has resulted in a great deal of interest in the unique Furin cleavage site of the Sars2 spike protein [1-3]. Furins are subtilisin-like peptidases that cleave at a predictable multibasic cleavage site typically consisting of either an RXRR or RXKR motif. Humans encode many Furin-like enzymes including human Furin and numerous proprotein convertase subtilisin/kexin type (PCSK) enzymes [4]. It has been shown that even within the RXRR motif Furins, there is substrate specificity depending on what the X is, and also some variation in preference for whether the terminal residues are RR or KR (e.g. PCSK3) [4-5]. However there is also residual activity of human Furin on RXXR sites in general (NEB: Furin), suggesting these enzymes are not absolutely specific. These cleavages are thought to occur either at the cell membrane, at the golgi, or in vesicles during subcellular trafficking [6]. There is some evidence that Sars2 infectivity is affected by the presence of the Sars2 Furin site (RRAR). In an African green monkey kidney epithelium cell line (Vero E6), loss of the Furin cleavage site is associated with improved infectivity [3]. However the direction of change owing to the presence/absence of this cleavage site in human cells and in more relevant tissues (like lung epithelium) is not clear. What is known is that such Furin sites are not unique to Sars2, but also found in MERS-CoV (RXVR), HCOV-OC43 (RSRR), and HKU1-CoV (RKRR) [7]. So it seems like Furin sites are maintained in many CoVs, despite a reduced efficiency of Furin site-containing Sars2 in Vero cells. Drosophila as a model for Furin evolution and specificity... The AMP genes I study using Drosophila are processed into multiple mature peptides by Furin cleavage. Drosophila melanogaster has three Furin enzymes, but knockdown of only Furin1 expression results in immune deficiency [8]. What is unique about the Drosophila model, in my experience investigating the evolution of Drosophila AMPs, is that different species appear to prefer different Furin cleavage sites even in evolutionarily conserved AMP genes. For instance, D. melanogaster AMPs typically encode an RXRR at their Furin cleavage sites, yet in D. neotestacea and other flies their Drosocin genes encodes RXVR (highlighted below). This is especially intriguing to me as RXVR is not even a known potential Furin motif; also note that RXVR is seen MERS-CoV. Other flies like Mosquitoes instead prefer an RXKR motif in their Attacin genes as best I can tell, yet moths like Bombyx mori and fruit flies (including D. neotestacea) encode using RXRR. It would be interesting to obtain a full picture of the cleavage sites used across multiple AMP genes and determine if there is a species-based preference in a subset of known Furin-cleaved genes. It would also be neat to analyze if there is selection on the residue directly upstream of these furin site as the Proline in front of the Sars2 site is quite the oddity, but is also found in e.g. Diptericin B of D. willistoni (see below). Drosocin (Dro) or Attacin C (AttC) have different Furin sites
Beyond insects, such Furin cleavage of immune genes appears to be extremely widely conserved across the tree of life. Big Defensins of mussels and clams also show an RXRR or RXKR preference difference in homologous Defensin genes in different species, where an RXTR is also seen in multiple clam genes, but never RXRR [9] (see below). In Atlantic Cod cathelicidins encode an RVRR cleavage site upstream of their C-terminal peptide product (see below with human cathelicidin) [11]. 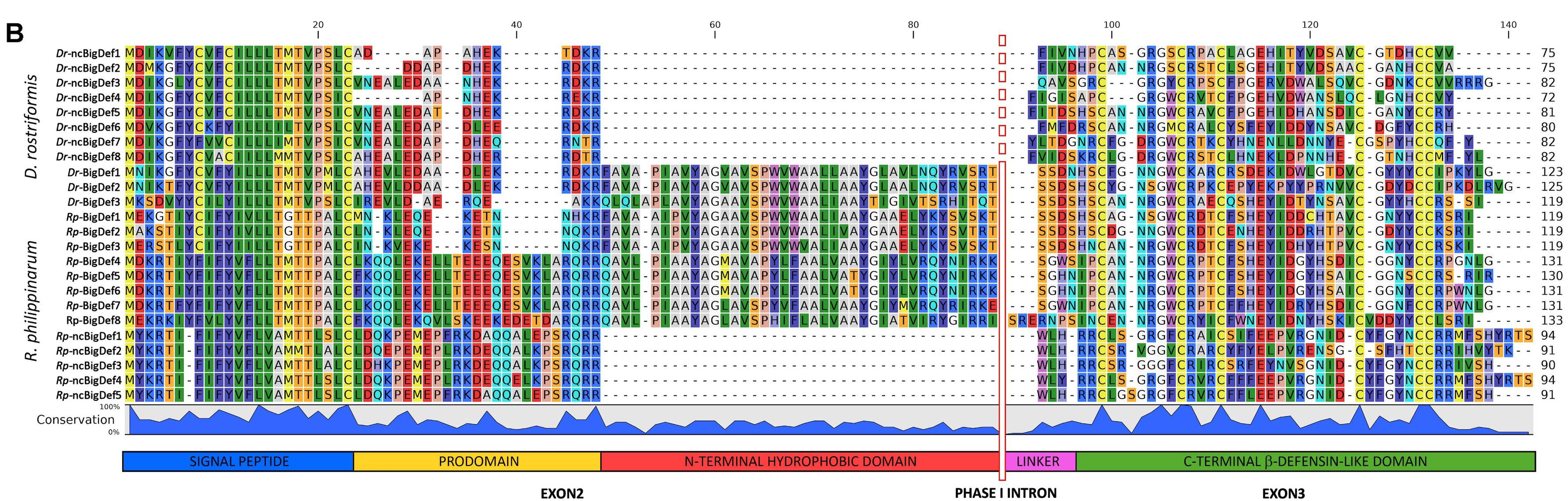 Figure 4B from Gerdol et al. [9] showing the RXKR, or RXRR sites of Defensins in the mussel Dreissena rostriformis and clam Ruditapes philippinarum. What's curious is that D. rostriformis never encodes an RXRR, while R. philippinarum encodes either RXKR (few) or RXRR (most). The putative Furin site is around residue 46 of the alignment. I don't see classic Furin sites in human AMP genes reviewed in Wang [10]. However I do recover dibasic sites (RR/KR) directly upstream of the mature AMP in certain human AMPs or AMP-like genes such as LL-37 (Cathelicidin) and Histatins (which produce many, many sub-peptides). Dibasic cleavage is typically performed by subtilisin-like proprotein convertases (like Furin) [12], suggesting that the cleavage leading to certain human AMPs also relies on Furin-like processing though perhaps not on Furin itself. Certainly cathelicidins of fish (including salmon, not shown here) display conserved RXRR Furin cleavage sites [11]. Furin / Dibasic sites Example mature peptides (underlined) >Hsap\CAMP (LL-37 peptide) MKTQRDGHSLGRWSLVLLLLGLVMPLAIIAQVLSYKEAVLRAIDGINQRSSDANLYRLLDLDPRPTMDGDPDTPKPVSFTVKETVCPRTTQQSPEDCDFKKDGLVKRCMGTVTLNQARGS FDISCDKDNKRFALLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES >Gadus morhua (Atlantic cod) Cathelicidin-1 MTTQMRLLCFAAVTLLAEAQMIPDPFIFPLKNFRPLLDQLRVETVYPEGVDLSTMSVRKMTFPAQELDCSQVNTSMPGQQCPLKENGKMMNCNFTLSYINQDADIQGFQFNCDAAIKEATLTRVRRSRSGRGSGKGGRGGSRGSSGSRGSKGPSGSRGSSGSRGSKGSRGGRSGRGSTIAGNGNRNNGGTRTA >Hsap\Histatin 3 (multiple mature peptides) MKFFVFALILALMLSMTGADSHAKRHHGYKRKFHEKHHSHRGYRSNYLYDN MKFFVFALILALMLSMTGADSHAKRHHGYKRKFHEKHHSHRGYRSNYLYDN MKFFVFALILALMLSMTGADSHAKRHHGYKRKFHEKHHSHRGYRSNYLYDN Applying this to the Sars2 Glycoprotein Furin-like enzymes are a core component of the production of immune effectors across the tree of life perhaps best characterized in invertebrates. This process might differ markedly between humans, clams, and flies, but the use of dibasic cleavage sites in AMP genes across all these species suggests that Furin-like enzymes have continued to be utilized in processing effectors of the innate immune response. Given the excitement around the Furin site of the Sars2 glycoprotein, the advantage of such a Furin site might be extremely species-specific depending on which Furin motif that species utilizes for its immune response. In the search for an animal model to replicate human Sars2, it may be useful to consider what Furin sites are associated with that species' immune response, and determine if there is a correlation between Furin site preference and symptom severity in existing animal model screens in say... cats and ferrets but not dogs [13]. It would also be very interesting to know if Furin site preference differs amongst bat species, and might correlate with Bat*CoV infectiousness dependent on CoV-Furin site presence and bat Furin site preference. Finally, suppressing Furin itself seems likely to affect many subcellular processes which seems bad for therapeutic potential, perhaps some therapeutic could be developed that inhibits the cleavage of Sars2 glycoprotein itself? This would rely on a demonstration that Furin cleavage of Sars2 enables virulence/infectivity in human cells, which remains unclear. Vaccines might also benefit from deleting this Furin site from the Sars2 virus to produce an attenuated virus that otherwise produces all the antigens needed to develop immunity to Sars2. In Conclusion I'm well out of my wheelhouse here, so by all means if you're reading and have anything to add/correct, please do! But Furin-like enzymes seem like a ubiquitous part of the innate immune response conserved throughout the tree of life, and at least in Vero cells, directly affect cytopathology [3]. References:
Comments are closed.
|
AuthorMark Archives
July 2024
Categories |
 RSS Feed
RSS Feed
